LLMs: App architecture + tings
I’m back and I’m still excited about LLM prompt engineering and orchestration. To recap, our team is building an EHR-embedded chat application. This will be delivered to users via a web embed (think HTML iframe). Here’s the evolving architecture map:
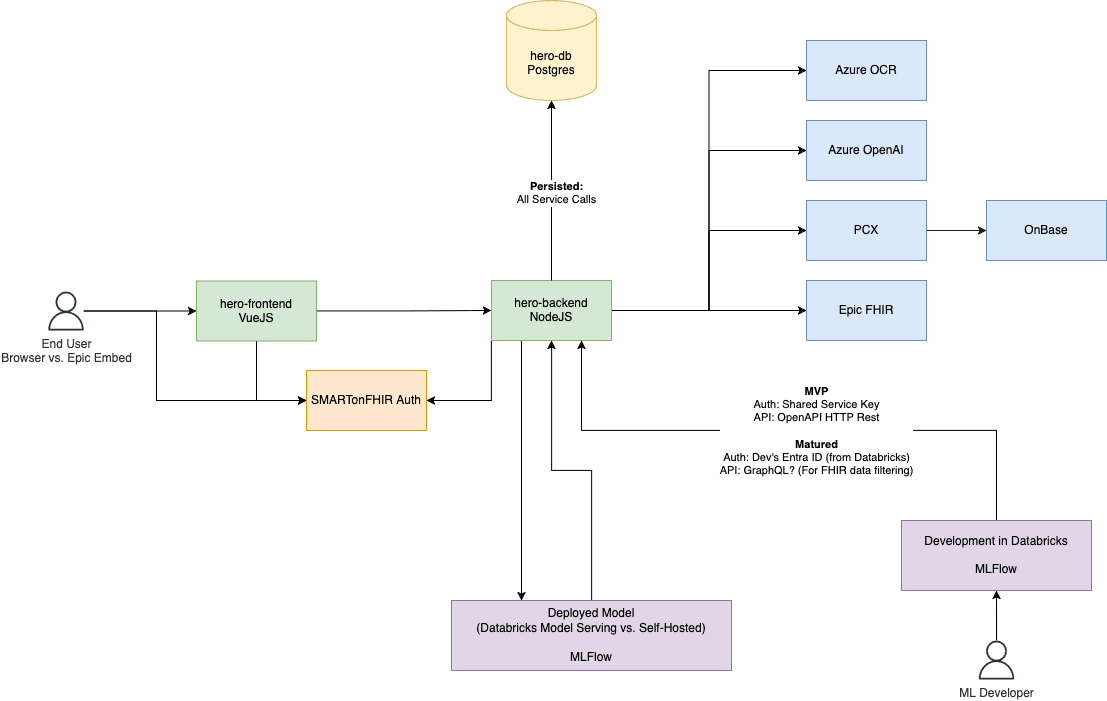
A couple interesting things to point out here:
- SMART on FHIR – we’ll be using SMART on FHIR as a single sign on method for the app, which is mostly follows OAuth conventions. When launched from within the EHR, the user’s EHR credentials will be used to grant access to the app and FHIR APIs used to retrieve clinical data. That data retrieval can take place on the frontend, backend, and in our LLM prompt flows, all with the same token. This means that any clinical data retrieved by the user or on behalf of the users are auditable as any other EHR activity.
- Decoupled application and "models" – by separating the base application (green boxes) from the flows (purple boxes) it serves to a user, we achieve a couple things:
- Non-web developers can contribute – I personally love the breadth of the whole stack, the frontend, backend, auth, persistence, Azure services, CI/CD, …all of it. But there’s also the converse interest, builders who excel at focusing more narrowly on deriving useful output from clinical data and in doing so dive way deeper into ML methods than I might. If we can agree on what input and output should look like using MLFlow, implementation can be left completely up to the ML builder.
- Supporting the ML model lifecycle – just like an application, a model deserves proper development tooling, telemetry (how many times has it been run, by whom, with which inputs/outputs, how long did it take), and a way to release version updates and roll them back if there are issues. MLFlow achieves these goals for us, and allows us to leverage deep integration with Databricks which our organization has already invested in. Finalized models can be deployed using model serving to fulfill requets originating from the frontend/backend..
- A bootstrapped dev environment – getting started building can be as simple as getting credentials to our workspace and internal APIs, cloning an example repo, and reading our internal API documentation.
As a testament to how useful this structure is for flow development, here’s an example of the kind of visual debugging we can do using MLFlow spans triggered by each step of our flow as well as all API calls it makes:
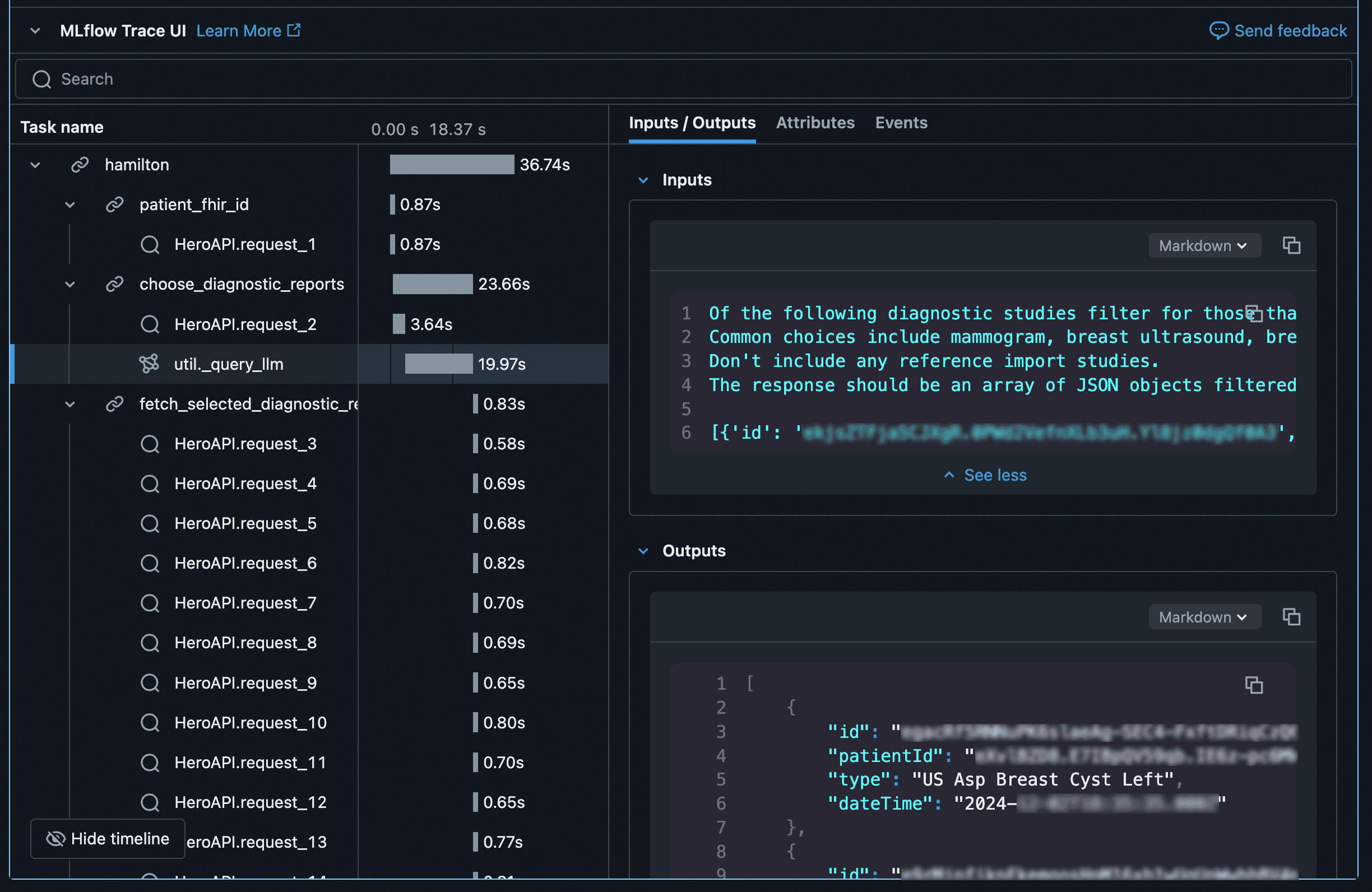
With this comes a confession, last post I promised I wouldn’t fall prey to premature optimization with regards to sync vs. async code in python. Well, I lied. I couldn’t bear the idea of making 10-15 HTTP calls in series while a user waits for the flow to complete. Fortunately, I was able to work with the Hamilton package maintainers to get what I needed to fashion an async-compatible MLFlow plugin which is what helped generate the output above. Kudos to the team for being so engaged with the open source community, I hope to contribute my async MLflow plugin back to the project once it’s hardened.
Last post I also mentioned clinical data deduplication as a next hurdle to overcome. In thinking through methods I learned that there are at least two types of deduplication.
A simple form is detecting when two strings match exactly. Using our clinical example of a breast cancer patient, an MRI-guided needle biopsy might generate two FHIR DiagnosticReport resources with different ids and finalization times, but the exact same text. In these cases, it’s trivial to cluster those that have exactly matching text. Remember this includes things like punctuation, capitalization, and spaces; computers don't play games.
Semantic deduplication is more nuanced and interesting. Here we generate embeddings, representations of word meanings and use statistical methods to evalute for overlap. For example, the strings “Example sentence.” and “This is an example sentence.” might be considered semantic duplicates depending on your threshold for overlap.
It didn’t take long for me to bump into an open-source package SemHash which makes it really easy and fast to do this kind of deduplication. It generates embeddings and then performs an approximate nearest neighbors sorting using CPU-optimized code that works just fine locally without a GPU. Here’s how we’d use it to implement the above example:
from semhash import SemHash
records = [
{"id": 1, "text": "Example sentence."},
{"id": 2, "text": "This is an example sentence"}
]
records = SemHash.from_records(records=records, columns=['text']).self_deduplicate()
print(records.duplicates)
Which would give us a result like this, the duplicate with it's similarity scoring:
[
DuplicateRecord(
record={'id': 2, 'text': 'This is an example sentence'},
exact=False,
duplicates=[
({'id': 1, 'text': 'Example sentence.'}, np.float32(0.9840234))
]
)
]
When we apply both approaches (exact and semantic deduplication) we’re able to achieve model output for our breast cancer flow like the following:
- 01/01/2085 - The right breast shows a partially cystic/necrotic and partially solid mass measuring up to X x Y x Z cm, containing a biopsy clip and corresponding to biopsy-proven malignancy. The mass is abutting the skin but shows no enhancement of the skin.
- MAMMO DIAGNOSTIC BILATERAL TOMOSYNTHESIS (BILATERAL ULTRASOUND IF NEEDED)
- Duplicate: MAMMO DIAGNOSTIC LEFT TOMOSYNTHESIS (ULTRASOUND IF NEEDED)
- Duplicate: MR BREAST W AND WO IV CONTRAST BILATERAL
- Duplicate: US BREAST BIOPSY LEFT
- Duplicate: US BREAST LEFT LIMITED
- Duplicate: US BREAST RIGHT LIMITED
In the above, the model has retrieved 6 DiagnosticReports and found that they all convey the same information, so why not just summarize the first? Is this something our EHR should just do for us? Maybe? Probably. Out of caution I am only running deduplication on studies within a 24 hour time period. I think this covers clinical workflows that cause these duplicate reports to be generated. Also, I wouldn’t want similar appearing studies months or years apart to be considered duplicates. Here’s what the code for this node of our DAG looks like:
async def deduplicate_diagnostic_reports(
fetch_selected_diagnostic_reports: List[dict],
time_window_seconds: int = 24 * 60 * 60,
semantic_dedup_threshold: float = 0.9
) -> dict[str, Any]:
"""
Run deduplication on reports that are within time_window_seconds of each other
"""
# Group reports together that are within time_window_seconds of each other
ungrouped_reports = fetch_selected_diagnostic_reports.copy()
report_groups = []
while len(ungrouped_reports) > 0:
new_group = [ungrouped_reports.pop(0)]
indexes_to_add = []
for i, ungrouped_report in enumerate(ungrouped_reports):
if abs(
datetime.fromisoformat(ungrouped_report['dateTime']).timestamp()
- datetime.fromisoformat(new_group[0]['dateTime']).timestamp()
) < time_window_seconds:
indexes_to_add.append(i)
for i in sorted(indexes_to_add, reverse=True):
new_group.append(ungrouped_reports.pop(i))
report_groups.append(new_group)
# Deduplicate each group with SemHash
deduplicated = []
semantic_duplicates = []
exact_duplicates = []
for group in report_groups:
report_dicts = []
for report in group:
joined_result_text = ' '.join([result['value'] for result in report['results']])
# Log and exclude exact duplicates
is_exact_duplicate = False
for rd in report_dicts:
if rd['text'] == joined_result_text:
exact_duplicates.append({ "retained_id": rd['id'], "excluded_id": report['id'] })
is_exact_duplicate = True
break
if not is_exact_duplicate:
report_dicts.append({ 'id': report['id'], 'text': joined_result_text })
dedup = SemHash.from_records(records=report_dicts, columns=['text']).self_deduplicate(threshold=semantic_dedup_threshold)
deduplicated.extend(dedup.deduplicated)
semantic_duplicates.extend(dedup.duplicates)
# Combine exact and semantic duplicates into a single lookup dict
duplicates: dict[str, List[DuplicateResource]] = {}
for sd in semantic_duplicates:
duplicates[sd.record['id']] = duplicates.get(sd.record['id'], [])
for duplicate in sd.duplicates:
duplicates[sd.record['id']].append(DuplicateResource(retained_id=sd.record['id'], excluded_id=duplicate[0]['id'], score=duplicate[1]))
for ed in exact_duplicates:
duplicates[ed['retained_id']] = duplicates.get(ed['retained_id'], [])
duplicates[ed['retained_id']].append(DuplicateResource(retained_id=ed['retained_id'], excluded_id=ed['excluded_id'], score=1.0))
return {
"report_groups": report_groups,
"semantic_duplicates": semantic_duplicates,
"exact_duplicates": exact_duplicates,
"deduplicated": deduplicated,
"duplicates": duplicates,
"duplicates_removed": len(fetch_selected_diagnostic_reports) - len(deduplicated)
}
As an aside, you'll see I excluded exact duplicates before using SemHash. It turns out the the version I started writing this node with had a bug that the fix hadn't been released for. I pinged the package maintainers and they took care of it wihtin a day. Isn't open source beautiful? 🤓