LLMs: To agent or not to agent
I’ve been excited about some new internal build our team has been exploring, an LLM-driven chat application that interfaces with our electronic health record (EHR). The premise is that clinicians and allied professionals might benefit from the capabilities of an LLM but need to maintain the privacy of a patient’s protected health information (PHI). Summarization is a common example with prompts like: “Draft a return-to-work letter after this patient’s cardiac surgery with rehab duration and precautions” or “Summarize this patient’s cardiac history including coronary anatomy, infarct pattern, and prior interventions based on prior imaging, procedures, and clinic notes.”
One approach is to create a chat “agent”, a chatbot that accepts conversational input, and leverages a suite of “tools” that perform retrieval augmented generation (RAG). Ideally, as the conversation progresses, the chatbot gets closer to what the user is looking for. I spent some time experimenting with LangGraph to implement a FHIR-based lookup tool and was impressed with what’s possible. The interesting technical magic about LLM tools is that input parameters are interpreted by the LLM and both steer the conversation and coalesce responses into parameters acceptable for the tool. This is where static typing and schema tools like zod and Pydantic absolutely shine – your function interface documentation (including natural language) serve as instructions for the LLM. Errors during validation can be fed back to the LLM and used to generate a response to the user asking for clarification. Anyway, here's an excerpt of my tool, that fetches demographics from FHIR based on a patient identifier from the conversation:
export const patientLookup = tool(
async (input, config: RunnableConfig) => {
const fhirRequest = config.configurable?.fhirRequest as ReturnType<typeof EpicService.prototype.fhirRequestFactory>
if (!fhirRequest) { throw new Error('Unable to make a FHIR request without fhirRequest function') }
try {
const patient = await fhirRequest({ endpoint: `Patient/?identifier=MRN|${input.patientId}`, method: 'GET' }) as fhir4.Bundle
return `Found patient, their name is: ${JSON.stringify(patient)}`
} catch (e) {
return `Failed to find patient ${input.patientId}: ${e}`
}
},
{
name: 'patientLookup',
description: 'Find a patient by id',
schema: z.object({
patientId: z.string()
})
}
)
However, after this experience I’m not certain an agentic flow is the approach I want to take. I feel that the path from parsing input from arbitrary instructions to arriving at useful output is too uncertain. I don’t think that clinicians want to invest time prompting a chatbot repeatedly to maybe get something useful. There’s also the possibility that your user might be disappointed that their prompt isn’t covered by the capabilities of the agent. Instead,
I hypothesize that a clinician wants something they can provide minimal parameters that will generate something useful within seconds to minutes. Any necessary parameters ought to be prepopulated from the available context within the patient’s chart where possible.
With that in mind, I am exploring what a non-agentic, directed acyclic graph (DAG) for summarization might look like. Stepping outside cardiovascular medicine, my specialty of practice, what if we wanted to generate an oncology history for a breast cancer patient? A typical oncology history for such a patient would include the timeline of presenting symptoms, relevant imaging (mammograms, MRI, ultrasound), biopsies and or surgical resections, pathology review, treatment cycles and response, and post treatment surveillance.
Taking a step back from LangGraph which I was excited about, but found challenging to take on from scratch, I bumped into hamilton – a simpler python framework for designing and running DAGs of any kind. Here’s what my first pass at this looks like, based only FHIR DiagnosticReports:
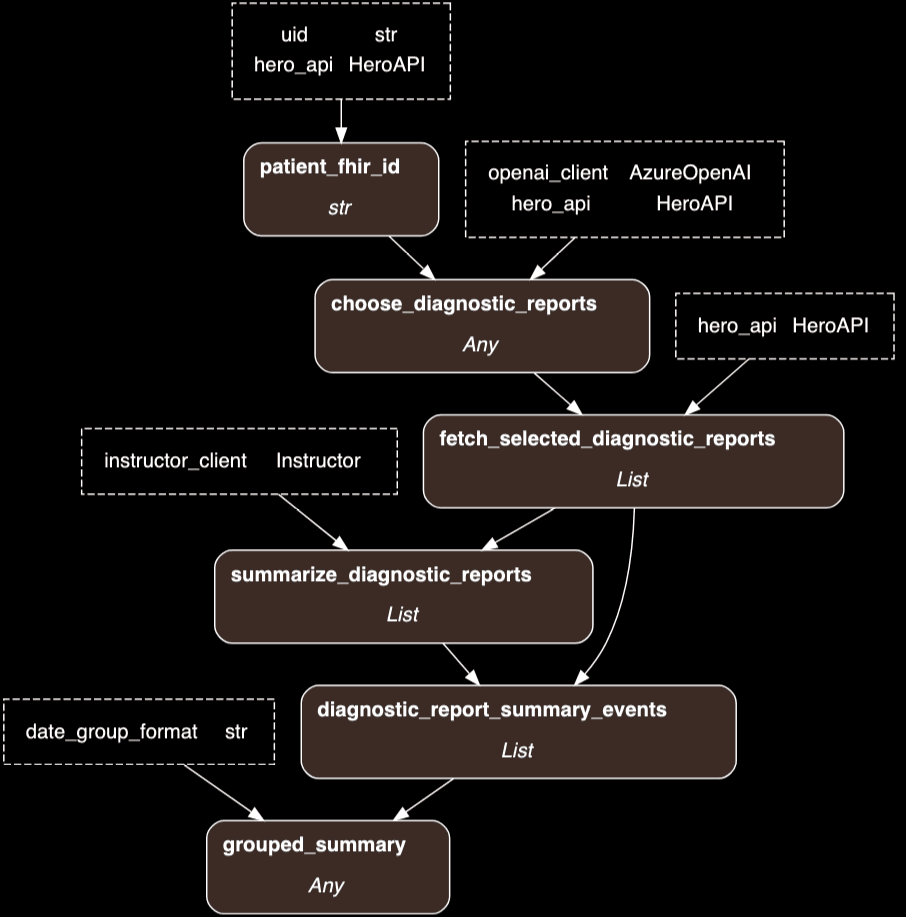
Here’s some hamilton code (elegantly, just a series of python functions with matching interfaces) to fetch and summarize relevant imaging. The code below takes a patient identifier (UID) as input, uses ChatGPT (via the OpenAI python client and Instructor) and an internal FHIR API called hero. The end result is a markdown summary that is returned to the frontend.
def patient_fhir_id(hero_api: HeroAPI, uid: str) -> str:
"""
Lookup patient
"""
patient = hero_api.request(endpoint='fhir/patient', params={"id": uid, "idType": "UID"})
fhir_id = patient['id']
return fhir_id
def choose_diagnostic_reports(openai_client: AzureOpenAI, hero_api: HeroAPI, patient_fhir_id: str) -> Any:
diagnostic_reports = hero_api.request(endpoint=f'fhir/patient/{patient_fhir_id}/DiagnosticReport')
prompt = f"""
Of the following diagnostic studies filter for those that may be related to breast cancer.
Common choices include mammogram, breast ultrasound, breast MRI, surgical pathology.
The response should be an array of JSON objects filtered from below:
{diagnostic_reports}
"""
response = _query_llm(openai_client, prompt)
return _parse_llm_json_response(response)
def fetch_selected_diagnostic_reports(hero_api: HeroAPI, choose_diagnostic_reports: dict) -> List[dict]:
report_ids = [r['id'] for r in choose_diagnostic_reports]
full_reports = []
for r in report_ids:
try:
full_reports.append(hero_api.request(endpoint=f'fhir/DiagnosticReport/{r}'))
except requests.exceptions.HTTPError as e:
logger.error(f'Skipping report {r} due to {e}')
full_reports = [r for r in full_reports if not isinstance(r, Exception)]
full_reports = sorted(full_reports, key=lambda x: x['dateTime'])
return full_reports
class DiagnosticStudySummary(BaseModel):
id: str
summary: str
normal: bool
event_type: Literal['imaging'] | Literal['pathology']
def summarize_diagnostic_reports(instructor_client: instructor.Instructor, fetch_selected_diagnostic_reports: List[dict]) -> List[DiagnosticStudySummary]:
"""
Summarize each report
"""
def generate_prompt(report: dict) -> str:
return f"""
Generate a summary of the following report.
The report may have more than one section but a single summary should be produced.
The summary should be concise and shold NOT include a description of the patient or their presentation.
Do not describe the absence of abnormalities.
Do not describe negative parts of the exam that were negative or absent of malignancy.
If the study was normal, the summary should say "Normal".
if the study was abnormal, the summary should describe the abnormality in a single sentence and include the following if available: measurements, BIRADS score.
The id value is: "{report['id']}"
{json.dumps(report['results'])}
"""
summaries = []
for r in fetch_selected_diagnostic_reports:
summaries.append(
_instuctor_llm(
instructor_client=instructor_client,
prompt=generate_prompt(r),
response_model=DiagnosticStudySummary
)
)
return summaries
class SummaryEvent(BaseModel):
id: str
source: str
datetime: datetime
event_type: Literal['imaging'] | Literal['pathology']
description: str
summary: str
class SummaryLine(BaseModel):
date: date
items: List[SummaryEvent]
def diagnostic_report_summary_events(
fetch_selected_diagnostic_reports: List[dict],
summarize_diagnostic_reports: List[DiagnosticStudySummary]
) -> List[SummaryEvent]:
summary_events = []
for report in fetch_selected_diagnostic_reports:
summarized = next((s for s in summarize_diagnostic_reports if s.id == report['id']), None)
summary_events.append(SummaryEvent(
id = report['id'],
source = 'DiagnosticReport',
datetime = report['dateTime'],
event_type = summarized.event_type,
description = report['type'],
summary = summarized.summary,
))
return summary_events
def grouped_summary(
diagnostic_report_summary_events: List[SummaryEvent],
date_group_format: str = '%B %Y',
) -> Any:
# collect all events
events = [
*diagnostic_report_summary_events
]
# Group by date_group_format
summary_lines = {}
for event in events:
grouper_key = event.datetime.strftime(date_group_format)
if grouper_key not in summary_lines:
summary_lines[grouper_key] = []
summary_lines[grouper_key].append(event)
# Generate markdown
markdown_lines = []
for group_title, events in summary_lines.items():
markdown_lines.append(f"# {group_title}")
sorted_events = sorted(events, key=lambda x: x.datetime)
for event in sorted_events:
markdown_lines.append(f"- [{event.description} {event.datetime.strftime('%m/%d/%Y')}]({event.id}): {event.summary}")
return '\n'.join(markdown_lines)
Next, I'll be working on deduplication - often the same information can appear in multiple places in the EHR. For example, an encounter for a breast biopsy might result in multiple reports, one for the ultrasound guidance, another for the biopsy itself, and another still for the pathology report. In the context of a timeline we'll want to distill these to a single relevant clinical event. Stay tuned.
From a technical perspective, one thing I’ve bumped into coding this in Python, that I’m familiar with solving in NodeJS is optimizing non-dependent asynchronous I/O bound (web API) calls. Examples of this above are the API calls to retrieve the FHIR DiagnosticReports or the ChatGPT summarization calls for each report. In JavaScript, I’d create an array of Promises and fire them off together with Promise.all. In Python, I need to contend with whether my code (and any underlying packages) should/can be parallelized with asyncio. Hamilton fortunately does have an asyncio-compatible driver, but it’s not the default and is missing some compatibility with features like MLFlow integration. After trying the asyncio path, I’ve reverted and want to focus my efforts on whether I can produce useful output first, even if it takes longer than I’d want in production. If I can solve for that, then I’ll reconsider asyncio or explore python multithreading.